Variants Structural Effects
Last update: 2023-01-20
Beside visual inspection of wild type and mutated 3D structures, MIZTLI can analyze and report some of the potentially damaging structural effects of an amino acid variant.
Detected Effects
MIZTLI tries to detect the following structural effects:
- Gain or loss of a disulfide bridge
- Introduction of a Proline in place of another amino acid in the protein core
- Steric clash between the variant amino acid and its surroundings
- Introduction of a hydrophilic amino acid in place of a hydrophobic one in the protein core
- Introduction of a charged amino acid in place of an uncharged one in the protein core
- Introduction of a positively, resp. negatively, charged amino acid in place of a negatively, resp. positively, charged one in the protein core
- Secondary structure altered
- Introduction of an uncharged amino acid in place of a charged one in the protein core
- Substitution of a Glycine in the protein core
- Loss of a buried hydrogen bond
- Loss of a buried salt bridge
- Variant amino acid buried, resp. exposed, to the protein surface whereas the wild type is exposed, resp. buried
- Introduction of a hydrophobic amino acid in place of an exposed hydrophilic one
- Substitution of a Glycine in a secondary structure bend
These potentially damaging structural effects are described in Ittisoponpisan, S., et al. (2019).
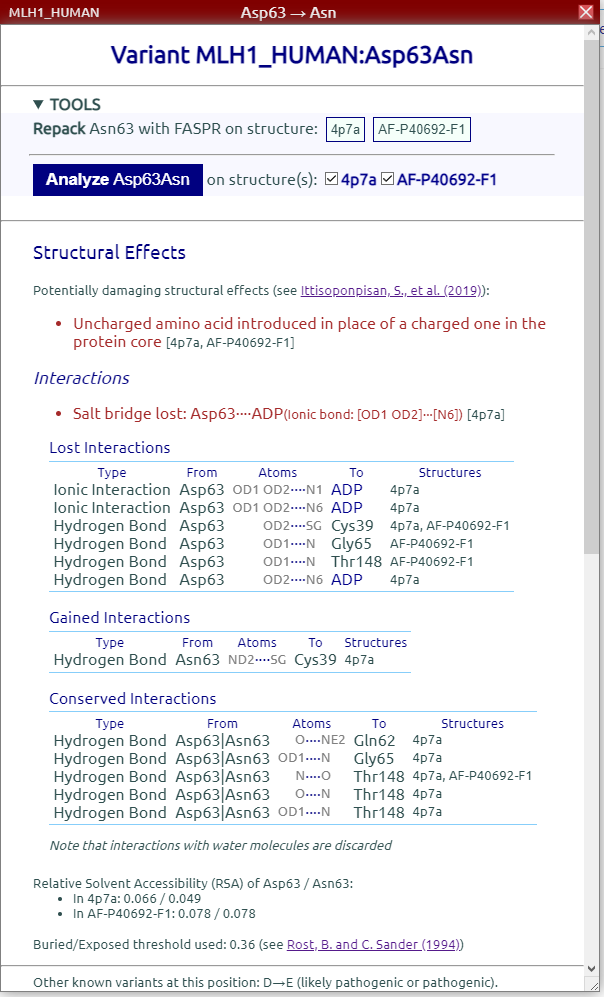
The structural effects report also includes details about lost, gained, and conserved non-covalent interactions within the protein and with ligands.
Procedure
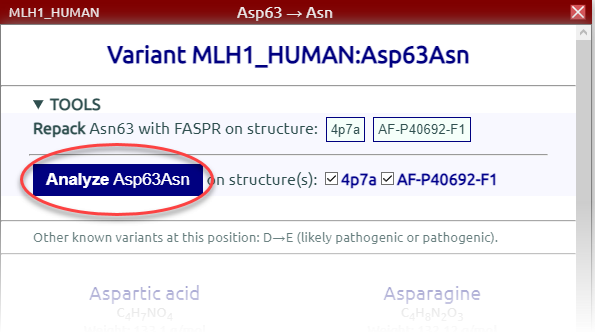
Create a variant (see the main Tutorial). As an example we will use MLH1_HUMAN:Asp63Asn. Click on "Analyze Asp63Asn":
After some computation time a Structural Effects report is displayed: